1 Introduction
1.1 A Subsection Sample
In recent years, electronic medical systems had become a universally accepted technology. Especially, the application of Electronic Medical Records (EMRs) provides great convenience for doctors in using and organizing medical information. However, the ever-expanding EMRs result in a tremendous working burden towards the clinical medics. To release the doctor from a heavy burden, researchers aim to automate this process by using deep learning models generating EMRs automatically from medical consultation dialogues.
To achieve this goal, we need to extract valid information from medical consultation dialogues in the first step, which is also the focus of this paper. Specifically, we propose a medical information extraction system based on an encoder-decoder framework to detect valuable medical items from the doctor-patient dialogues.
Different from existing models[1,2,3], for example, MIE[4] only considers the interactive information of the dialogue within the window, our model also considers the global information from different windows, where semantic features of the model are enhanced and the model can take advantage of distant medical information in the scope of the entire dialogue. In the experiment, we use a public dataset MIE[4] to compare our model with baseline models. The experimental results show that our model improves 1.8% and 0.85% F1 scores on window-level and dialogue-level compared with the existing state-of-the-art model, which verifies the effectiveness of our model.
In the following, we will introduce the representative works in this field in Section 2, and our model design in Section 3, and our detailed experimental setup and result analysis in Section 4. Finally, we will summarize the full paper in Section 5.
2 Related Work
Extracting information from medical discourse text has just recently become possible. [5] presented a pipeline with five modules to generate EMR. We are primarily interested in the knowledge extraction module, which blends rule-based and supervised machine learning algorithms. [1] used 186 symptoms and three pre-defined statuses to extract symptoms and their respective statuses. They also introduced a span-attribute tagging approach, which predicted the range of symptoms reported before using context characteristics to predict the symptom name and condition. [2] annotated Chinese online medical discussions with the BIO (beginning, inner, or other) schema. However, they just marked the symptoms as entity types and did not examine the state, which is problematic. The most recent work is MIE [4], which has a more extensive annotation schema with 4 categories, 71 items, and 5 statuses, and the suggested pipeline architecture consists of 4 sub-modules to iteratively classify the entire labels.
3 Model
As shown in Figure 1, our model consists of three parts. The first part is called window encoder, which can convert the original input text into a fixed dimension feature matrix; The second part is a global information aggregation module named global information aggregator, which integrates the interactive information between different dialogue windows. The third part is the medical label predictor, which uses a binary classifier to iteratively predict each candidate label item across the whole label set.
There are three model inputs, which include the raw text of the current window, the raw text of the following windows and the candidate label. Among them, each window contains two sentences, representing the dialogue information between doctors and patients, respectively.
Figure 1. Model Overview
3.1 Window encoder
In this module, we use \( {x_{i}} \) and \( {l_{i}} \) to indicate every window and label individually. At first, we use a word embedding layer to transfer the text into matrixes containing semantic information. Then we map \( {x_{i}} \) and \( {l_{i}} \) to a higher dimensional matrix \( {h_{i}} \) and \( {g_{i}} \) by using a Bi-directional LSTM network[6,7].
\( {h_{i}}=BiLSTM({x_{i}}) \) (1)
\( {g_{i}}=BiLSTM({l_{i}}) \) (2)
3.2 Global information aggregator
In this module, the output of the window encoder \( {h_{i}} \) in section 3.1, will be concatenated to the feature matrix of the following windows \( {\lbrace h_{i+1}},{h_{i+2}},…,{h_{t}}\rbrace \) , in which the t represents the number of following windows in the entire dialogue. However, not all following windows can bring valid semantic information which benefits the model performance. On the contrary, for those invalid windows, their semantic information will bring extra noise to the medical information prediction of the current window, which will potentially deteriorate the final performance of the model. Therefore, instead of considering all feature matrixes of the following windows, we only select the most informative window as our global feature, so as to assist in the extraction of medical information from the current window. The reason for ignoring the dialogue history information is that, according to our observation of the dataset, the information decision of current window is mainly determined by the following windows, and not affected by the previous windows.
We adopt a dynamic attention mechanism[8] to achieve the above effect. Different from the conventional attention mechanism, the proposed dynamic attention is changing constantly on each window, that is, the number of windows below each window is different. In other words, each window has a different number of the following windows. For example, there are 5 windows in total, so the second window has 3 following windows; the 4th one has only 1 following window and the last window has no following window. In this case, when calculating the attention weight, the number of attention scores considered by our attention mechanism is gradually changing, where it is fixed in the traditional attention mechanism. Therefore, we call our method the dynamic attention mechanism.
Concretely, given current window \( {h_{i}} \) and its corresponding lower window \( {\lbrace h_{i+1}},{h_{i+2}},…,{h_{t}}\rbrace \) , we use \( {h_{i}} \) as the query of attention to calculating attention score \( {a_{ij}} \) of all following windows.
\( {a_{ij}}=\frac{{h_{i}}∙h_{j}^{T}}{\sqrt[]{{d_{x}}}} \) (3)
Where \( {d_{x}} \) represents the dimension of the word vector. T represents matrix transpose operation. And \( {a_{ij}} \) indicates the attention score of the following window \( {h_{j}} \) to the current window \( {h_{i}} \) . Then we calculate the attention weight \( {w_{ij}} \) of each following window.
\( {w_{ij}}=\frac{{℮^{{a_{ij}}}}}{\sum _{k=i+1}^{t}{℮^{{a_{ik}}}}} \) (4)
After that, \( {h_{d}} \) , the lower window which has the highest attention weight[9], will be selected as our global information to enhance the semantic information of the current window. Thus, we get \( h_{i}^{ \prime } \) , a new current window integrating global information.
\( h_{i}^{ \prime }=λ∙{h_{i}}+(1-λ)∙ {h_{d}} \) (5)
In which, \( λ \) is an untrainable hyper-parameter used to balance \( {h_{i}} \) and \( {h_{d}} \) .
4 Experiment
4.1 Dataset
Our data comes from Chunyu-Doctor, a Chinese online medical consultation platform[10]. Every dialogue between doctor and patient is completed on the internet and stored in text format. Specifically, MIE dataset includes 4 categories: symptom, surgery, test and other information. These 4 categories can be further divided into 72 items and each item has a status: negative, positive or unknown. MIE is a medical dialogue information extraction dataset that can be summed up into these characteristics: (1) The annotation of each medical information is coarse-grained. In other words, instead of annotating the entity itself, MIE just annotates the sentences that contain the specific information. Therefore, the MIE task is identified with a sentence classification task. (2) Single specific medical information, category label, can occur multiple times in the MIE dataset. In terms of statistical information, MIE contains 1120 doctor-patient dialogues and 46151 annotated labels. We use the same division with the original MIE which is divided into 800/160/160 respectively used for train/develop/test sets. According to the information provided by MIE, few windows do not contain any medical information and we use None to annotate these uninformative windows individually.
4.2 Experimental settings/model Details
To ensure the fairness of comparison, we keep our parameter settings consistent with our baseline model. Concretely, we use 300-dimensionality Skip Gram[11] as our model’s word embedding and we get our embedding by pre-training on MIE data. In our model, the sizes of hidden layers of both LSTM and fully-connected layers are set to 400. Meanwhile, we perform a dropout operation, whose probability is 0.3, before the output of each layer to prevent the potential overfitting issue. We select Adam optimizer[12] and use F1 score of testing set to carry out the early stop.
4.3 Experimental Results
The same with the baseline model, we examine our model at window-level and dialogue-level. We adopt Precision, recall and F1 score as our evaluation metrics. Concretely, window-level represents that we calculate those three metrics for each window separately, then add and average them as the final result; Dialogue-level means we evaluate the whole dialogue as an entirety, while the duplicate labels will be excluded.
Table 1. Experimental results of MIE task with window-level and dialogue-level
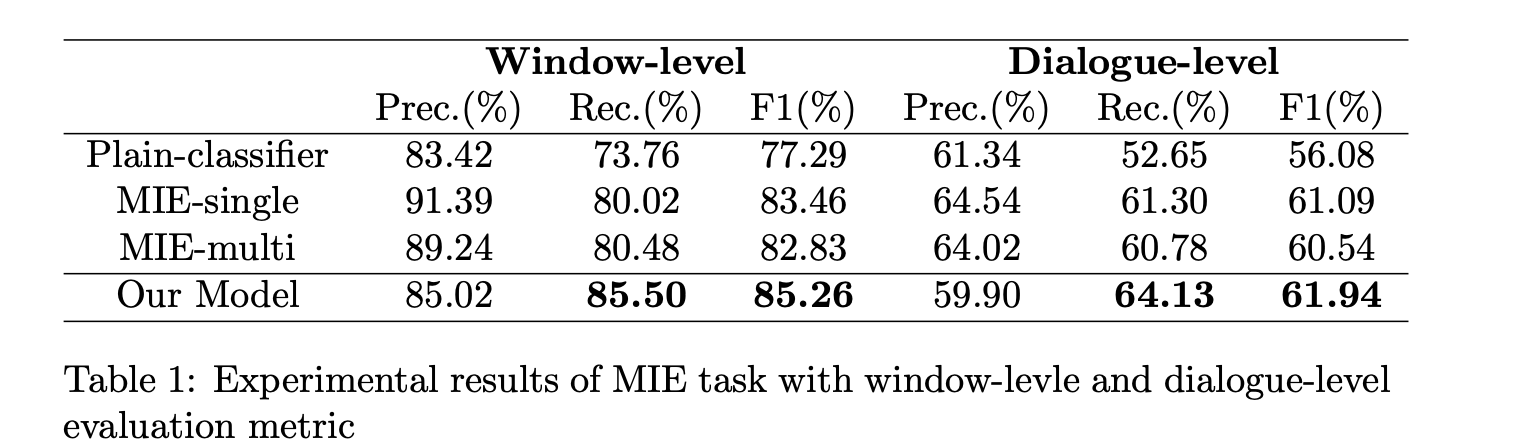
As shown in Table 1, there is no doubt that Plain-classifier gets the lowest result because no interactive information between sentences is considered. Besides, we notice that both MIE-single and MIE-multi get similar results which far surpass Plain-classifier in window-level and dialogue-level. Speaking of ours, we transcend the baseline model of 4.02% recall and 2.34% F1 score in the window-level, 3.35% recall and 1.40% F1 score in the dialogue-level. All of these validate the effectiveness of our model.
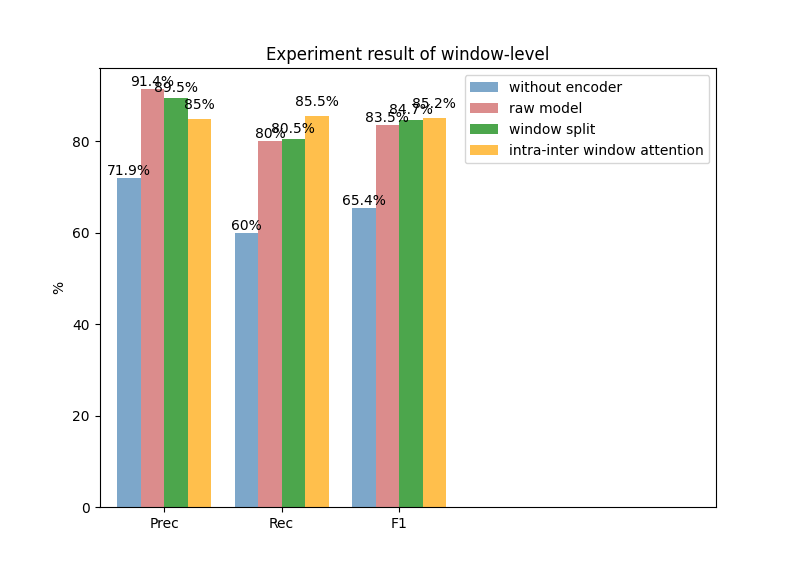
(a)
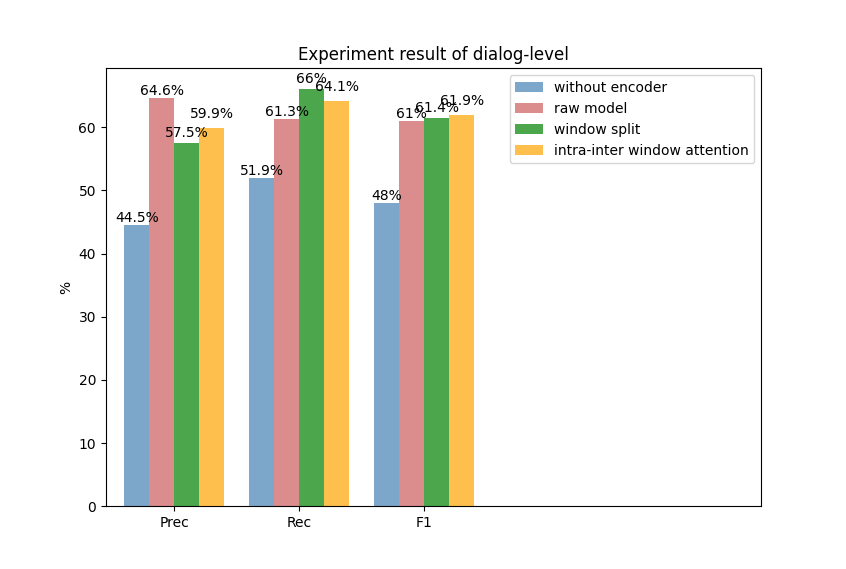
(b)
Fig. 2. Experiment results of window level (a) and dialogue level (b)
We also conduct a quantitative experiment to measure the effectiveness of the various model components in the MIE task. As shown in Figure 2, raw model is similar to MIE-multi, which has been used as our compared baseline. We firstly remove the model encoder parts, which represents we only obtain the word embedding representations before our model classification layer, we can notice the performances drop dramatically in both window and dialogue levels. Besides, we can see from Figure 2 that the model performance boosts 1.27% and 0.38% F1 scores on window level and dialogue level if we conduct a more reasonable window split strategy, that is set the window size to 2. Finally, according to integrating the global information, we can notice the model performance improves 0.51% F1 and 0.47% F1 scores, which also proves our model’s effectiveness.
5 Conclusion
In this paper, we built a clinical information extraction system, and our model effectively and completely leverages important context from the following windows in order to better capture state updates. Experiments in MIE tasks have shown that our model constantly improved performance and outperformed the baselines. This indicates our method can be a promising solution for real-world clinical practice.



