1. Introduction
The rise of drug-resistant bacterial strains poses a significant threat to global public health, challenging the efficacy of antimicrobial therapies and complicating the management of infectious diseases. Despite the remarkable progress made in medical science and technology, the evolution and dissemination of resistance mechanisms persist as formidable hurdles in the fight against microbial pathogens. In this landscape of escalating resistance, molecular epidemiology emerges as a beacon of hope, offering insights into the genetic underpinnings, transmission dynamics, and evolutionary trajectories of drug resistance. Molecular epidemiology, a multidisciplinary field at the intersection of microbiology, genetics, and epidemiology, harnesses an array of sophisticated molecular biology techniques to unravel the complexities of drug resistance. Techniques such as whole-genome sequencing, polymerase chain reaction (PCR), and pulsed-field gel electrophoresis (PFGE) empower researchers to probe into the genetic landscape of bacterial pathogens with unprecedented precision and depth. By deciphering the genetic makeup of drug-resistant strains, molecular epidemiologists can discern the molecular mechanisms underpinning resistance phenotypes and trace their origins and dissemination pathways. Central to the study of drug resistance is the exploration of genetic diversity within bacterial populations. Through comparative genomics analyses, researchers elucidate the myriad genetic variations that contribute to resistance, shedding light on the evolutionary forces driving their emergence and spread. Moreover, molecular epidemiology provides invaluable insights into the transmission dynamics of drug-resistant pathogens, both within healthcare settings and broader community environments. By employing molecular typing methodologies such as multilocus sequence typing (MLST) and SNP analysis, researchers can delineate strain lineages and trace transmission routes, facilitating the development of targeted infection control measures and public health interventions. Furthermore, molecular epidemiology endeavors to unravel the complex interplay of factors influencing the evolution of drug resistance. Antibiotic usage patterns, horizontal gene transfer mechanisms, and host-pathogen interactions all contribute to the selective pressures shaping the evolution of resistance phenotypes [1]. Through rigorous investigation and quantitative analysis, researchers aim to identify the key drivers of resistance emergence and dissemination, paving the way for the development of innovative therapeutic strategies and antimicrobial stewardship initiatives. In this review, we delve into the multifaceted landscape of drug resistance through the lens of molecular epidemiology, exploring the genetic diversity, transmission dynamics, and evolutionary determinants of resistance mechanisms. By synthesizing current research findings and insights, we aim to provide a comprehensive overview of the state of knowledge in this critical field, highlighting avenues for future research and intervention strategies to combat the global threat of drug-resistant infections.
2. Genotype Analysis of Drug-Resistant Bacterial Strains
2.1. Genetic Diversity and Evolutionary Relationships
Harnessing the power of cutting-edge molecular biology techniques, such as whole-genome sequencing (WGS) and polymerase chain reaction (PCR), researchers embark on a journey of comprehensive genotype analysis of drug-resistant bacterial strains. These sophisticated methodologies not only facilitate the sequencing of entire bacterial genomes but also enable targeted examination of specific genetic loci within bacterial isolates [2]. By leveraging comparative genomics approaches, researchers embark on the intricate task of reconstructing phylogenetic trees, shedding light on the complex genetic diversity and evolutionary relationships among disparate strains. Through meticulous phylogenetic analysis, researchers untangle the ancestral origins of drug resistance genes and decipher their dissemination patterns within bacterial populations. Moreover, genomic epidemiology serves as a beacon, illuminating the emergence of multidrug-resistant clones and delineating the adaptive evolution of resistance mechanisms over temporal scales. This amalgamation of cutting-edge molecular biology techniques and insightful genomic epidemiology opens new vistas for understanding the dynamic interplay between genetic diversity, evolutionary trajectories, and drug resistance in bacterial populations.
2.2. Genomic Characterization of Resistance Mechanisms
The genotype analysis of drug-resistant bacterial strains serves as the bedrock for discerning and delineating resistance mechanisms at the molecular level. Through meticulous examination of genetic determinants such as resistance genes, mutations, and mobile genetic elements, researchers embark on a journey to unveil the intricate molecular underpinnings of resistance phenotypes. Genomic investigations shed light on the diverse spectrum of resistance mechanisms prevalent across a myriad of bacterial species, underscoring the pivotal role of horizontal gene transfer in the dissemination of resistance genes. Moreover, genomic surveillance emerges as a beacon, facilitating the timely identification of novel resistance mechanisms and emerging threats [3]. This proactive approach not only informs the development of innovative therapeutics but also guides the implementation of antimicrobial stewardship initiatives aimed at preserving the efficacy of antimicrobial agents. Through the lens of genomic characterization, researchers gain invaluable insights into the dynamic landscape of resistance mechanisms, paving the way for targeted interventions and precision medicine approaches in the battle against drug-resistant infections.
2.3. Molecular Typing for Epidemiological Investigation
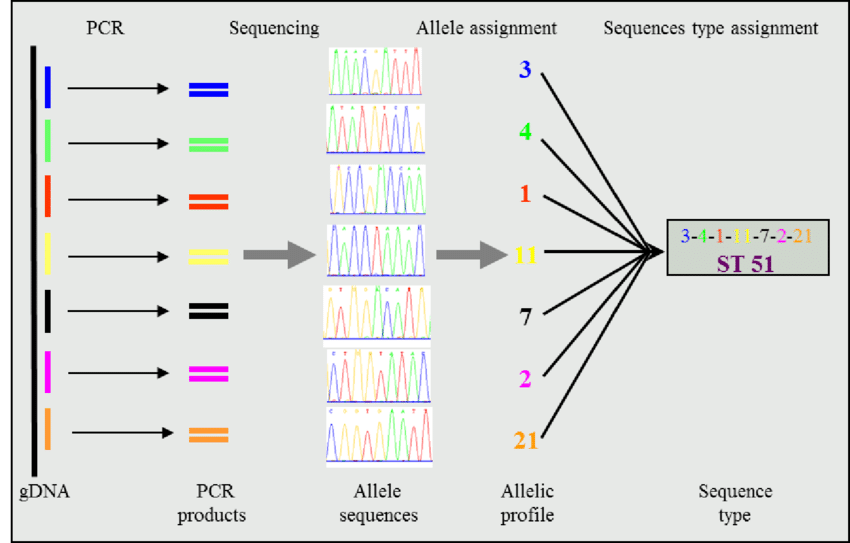
Figure 1. Scheme for multilocus sequence typing (Source: ResearchGate.com)
Complementing genotype analysis, molecular biology methodologies are harnessed for molecular typing of bacterial strains, thereby bolstering epidemiological investigations. Techniques including multilocus sequence typing (MLST)(as shown in Figure 1), pulsed-field gel electrophoresis (PFGE), and single-nucleotide polymorphism (SNP) analysis enable the delineation of strain lineages and the tracing of transmission routes. Molecular typing data furnish crucial insights for outbreak detection and containment efforts, facilitating the implementation of targeted infection control measures and public health interventions. Furthermore, molecular epidemiology studies offer insights into transmission dynamics within healthcare settings and communities, contributing to the surveillance of drug-resistant infections and the mitigation of nosocomial transmission [4].
3. Transmission Dynamics of Drug Resistance
3.1. Spread within Healthcare Facilities
The dissemination of drug-resistant bacterial strains within healthcare facilities constitutes a significant challenge in the realm of infection control and patient safety. Through the lens of molecular epidemiology investigations, critical insights into the complex dynamics of transmission within hospitals, clinics, and long-term care facilities have been unearthed. Harnessing the power of molecular typing methodologies such as pulsed-field gel electrophoresis (PFGE) in Figure 2 and single-nucleotide polymorphism (SNP) analysis, researchers meticulously unravel the intricate web of inter-patient and environmental spread of resistant strains. These comprehensive studies unveil the pervasive role of nosocomial transmission in perpetuating the propagation of drug resistance and instigating healthcare-associated outbreaks [5]. A nuanced comprehension of transmission dynamics within healthcare settings serves as the cornerstone for crafting tailored infection control measures and curtailing the dissemination of resistant pathogens. Through vigilant surveillance and targeted interventions, healthcare facilities can mitigate the risk posed by drug-resistant bacterial strains, safeguarding both patient well-being and public health.
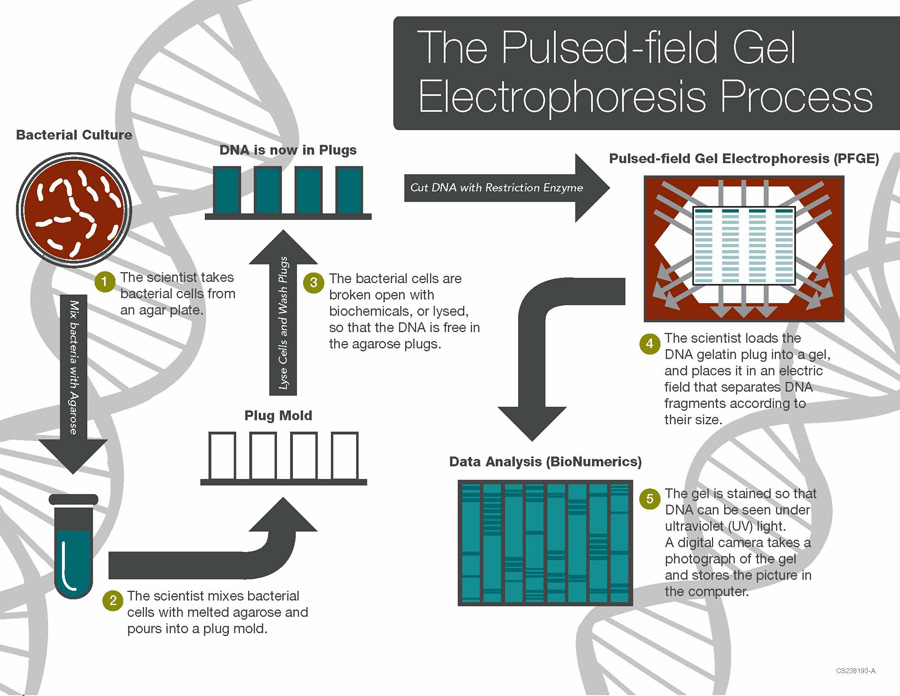
Figure 2. The Pulse Field Gel Electrophoresis (PFGE) Process (Source:biologyexams4u.com)
3.2. Community Transmission and One Health Perspective
Beyond healthcare facilities, drug-resistant bacterial strains exhibit propensity for dissemination within communities through multifarious avenues. Molecular epidemiology endeavors underscore the imperative of adopting a One Health approach, which acknowledges the interconnectedness of human health, animal health, and environmental factors, in elucidating the transmission dynamics of drug resistance. By amalgamating genomic datasets from human, animal, and environmental reservoirs, researchers illuminate the intricate transmission pathways of resistant strains across diverse ecosystems [6]. These investigations pinpoint agricultural antibiotic usage, livestock management practices, and environmental contamination as pivotal drivers of resistance dissemination. Embracing a One Health perspective is pivotal for formulating holistic interventions aimed at mitigating the spread of resistant pathogens and safeguarding antimicrobial efficacy.
3.3. International Spread and Global Surveillance
The transboundary dissemination of drug-resistant bacterial strains via international travel and trade poses profound implications for global health security. Molecular epidemiology research assumes a pivotal role in bolstering international surveillance endeavors aimed at monitoring the global spread of resistance. By fostering data sharing and cross-border collaboration, researchers discern the migratory patterns of resistant strains and anticipate emerging threats. Multilateral initiatives such as the Global Antimicrobial Resistance Surveillance System (GLASS) serve as conduits for information exchange and harmonized surveillance efforts across nations and regions [7]. These collaborative undertakings facilitate early detection of international outbreaks and inform the formulation of concerted response strategies. Augmenting global surveillance frameworks is imperative for curtailing the transnational dissemination of drug resistance and upholding the efficacy of antimicrobial agents worldwide.
4. Factors Influencing the Evolution of Drug Resistance
4.1. Antibiotic Usage and Selective Pressure
The pervasive utilization of antibiotics constitutes a primary driver in shaping the evolution and dissemination of drug-resistant bacterial strains. Antibiotics exert selective pressure on bacterial populations, favoring the survival and proliferation of resistant mutants. Molecular epidemiological investigations have delineated a clear association between antibiotic exposure and the emergence of resistance, underscoring the imperative of judicious antibiotic stewardship practices [8]. Through meticulous analysis of antibiotic consumption patterns vis-à-vis the prevalence of resistance, researchers discern patterns of resistance emergence and identify populations at heightened risk. Table 1 illustrates the relationship between antibiotic usage and the prevalence of resistance among bacterial strains [9].
Table 1. Relationship Between Antibiotic Usage and Prevalence of Resistance
Antibiotic | Usage (kg per capita) | Prevalence of Resistance (%) |
Penicillin | 15 | 30 |
Cephalosporins | 10 | 25 |
Macrolides | 8 | 20 |
Fluoroquinolones | 5 | 15 |
Tetracyclines | 7 | 18 |
4.2. Horizontal Gene Transfer Mechanisms
Horizontal gene transfer (HGT) mechanisms serve as crucial conduits for the horizontal dissemination of antibiotic resistance genes among diverse bacterial species, thereby contributing significantly to the evolution of drug resistance. These mechanisms, including conjugation, transformation, and transduction, facilitate the exchange of genetic material between bacterial cells, enabling the acquisition and propagation of resistance determinants. Through meticulous molecular epidemiological investigations, researchers have unveiled the multifaceted role of HGT in driving the evolutionary dynamics of drug resistance [10]. Conjugation, a process wherein genetic material is transferred through direct cell-to-cell contact via conjugative plasmids or other mobile genetic elements, plays a pivotal role in disseminating resistance genes among bacterial populations. Transformation involves the uptake and integration of exogenous DNA by recipient cells, often mediated by competence factors or environmental cues. Transduction, mediated by bacteriophages, enables the transfer of bacterial DNA between cells, facilitating the spread of resistance determinants. The intricate mechanisms underpinning the mobilization and transfer of resistance genes across bacterial populations underscore the importance of understanding HGT dynamics in combating drug resistance [11]. By unraveling the intricacies of HGT mechanisms, researchers can anticipate the emergence of novel resistance phenotypes and devise targeted strategies to disrupt transmission routes, thereby mitigating the spread of drug-resistant pathogens.
4.3. Host-Pathogen Interactions
The interplay between bacterial pathogens and their hosts constitutes a pivotal determinant in shaping the evolution of drug resistance. Bacterial pathogens adeptly navigate host immune responses and environmental pressures, engendering the selection of resistant variants endowed with heightened survival fitness. Molecular epidemiology studies offer invaluable insights into the genetic underpinnings of host adaptation and immune evasion, unraveling the intricate dynamics of drug resistance evolution. By scrutinizing the intricate interplay between bacterial pathogens and their hosts, researchers identify novel therapeutic targets and conceptualize innovative treatment modalities to combat drug-resistant infections [12].
5. Conclusion
In summary, the field of molecular epidemiology stands as a cornerstone in the ongoing battle against drug-resistant bacterial strains, offering profound insights into the genetic underpinnings, transmission dynamics, and evolutionary trajectories of resistance mechanisms. Through the meticulous analysis of genetic diversity, researchers have unveiled the intricate web of genetic variations that contribute to resistance phenotypes, shedding light on the adaptive evolution of drug-resistant strains. Moreover, molecular epidemiology has provided critical insights into the transmission dynamics of drug-resistant pathogens, both within healthcare facilities and broader community environments, enabling the development of targeted infection control measures and public health interventions. Looking ahead, it is imperative to adopt a holistic approach to combat drug resistance, embracing the principles of One Health and recognizing the interconnectedness of human health, animal health, and environmental factors. By integrating genomic data from diverse sources, including humans, animals, and the environment, researchers can gain a comprehensive understanding of the transmission pathways of drug-resistant strains across ecosystems. Furthermore, strengthening global surveillance efforts and fostering collaboration across borders are essential for monitoring the spread of resistance on a global scale and facilitating timely interventions. As we confront the formidable challenge of drug resistance, collaboration and concerted action are paramount. By leveraging the insights gleaned from molecular epidemiology research, policymakers, healthcare professionals, and researchers can develop evidence-based strategies to mitigate the spread of drug resistance and preserve the efficacy of antimicrobial agents. Through collective efforts and sustained investment in research and innovation, we can overcome the threat posed by drug-resistant pathogens and safeguard public health for generations to come.



